Non-equilibrium Polysome Dynamics
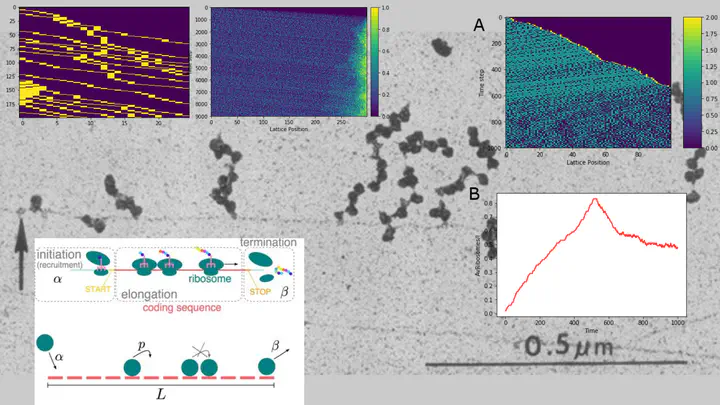 Photo by Gonza10V
Photo by Gonza10VIn bacteria transcription and translation are closely related and consists of a complex system of mechanical and chemical interactions that lead to gene expression. A common approach to model translation is trough ordinary differential equations (ODE), but in order to consider the complexity of this phenomenon has been modelled in terms of totally asymmetric simple exclusion process (TASEP) that came from non-equilibrium statistical physics. This model has interesting properties like the formation of three regimes: low density (LD), high density (HD) and maximum current (MC). Also has two transitions: from LD or HD to MC is lineal and from LD to HD is non-lineal. We write a python code that modelled translation like a TASEP and test the impact of opposing initial condition when the RNA start being translated, empty or full of ribosomes and found that the flux in HD regime with full start is similar to the flux in LD regime with empty start and the flux in LD regime with full start is similar to the flux in HD regime with empty start. Since the initial condition change the behaviour of translation, we modelled the coupling of transcription and translation with a two particles TASEP, ribosomes and an RNA polymerase (RNAP), we found that transcription and in special the RNAP hoping rate and pauses sets the initial conditions and can influence translation. Such a cooperative mechanism can improve RNA stability and open new ways to tune gene expression.