Build planning standardization
 Representation diagram
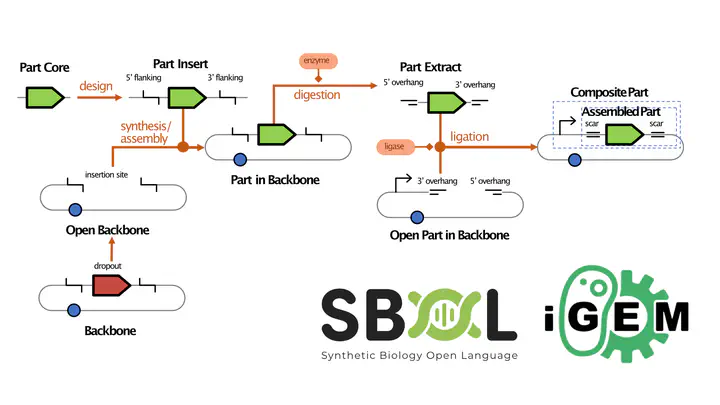
Representation diagramOne of the most common tasks in synthetic biology is building genetic constructs by assembling smaller parts. Despite this commonality, however, there is often much confusion when practitioners communicate about parts, sequences, and build plans. Parts often go through many stages during a build process, each with a different sequence. For example, a fragment of DNA may be synthesized as an insert into a vector backbone, then digested out of that backbone and assembled together with other fragments to produce a final construct. At present, without a shared standard for describing build plans, it is often difficult to tell which stage a given sequence is describing, leading to frequent confusion, errors, difficulty sharing information, and waste. We address this problem with a standard vocabulary for describing build plans, which we have further mapped into a concrete representation using the SBOL 3 standard. Specifically, we target representation of assembly based on digestion and ligation, supporting at least BioBricks Assembly and Type IIS assemblies like GoldenGate, MoClo, and GoldenBraid. The resulting vocabulary should be useful to practitioners no matter what tools or representations they may be using, while representation in SBOL 3 provides full details for use by software tool builders.